A new paper from Matt Hepworth and colleagues reveals a potential avenue for treating inflammatory disease.
Meanwhile, bacterial defence systems explored in Tim Blower’s latest paper could be translated into tools for studying human epigenome.
A new Nature Immunology paper from Lister Prize Fellow Dr Matt Hepworth offers insights into the activity of innate lymphoid cells and may prove useful in understanding and treating inflammatory diseases.

Innate lymphoid cells (ILCs) are an innate immune cell type discovered several years ago. They are mainly found in barrier tissues such as the intestinal tract, the lining of the lungs and the skin to form part of the first line of defence against bacteria or viruses.
In inflammatory diseases, such as inflammatory bowel disease, Crohn’s disease, asthma, and psoriasis, these cells become hyper-activated and appear to be one of the causes of inflammation in these diseases. A better understanding of how transcriptional networks support the function of ILCs could translate into therapeutic use to offer treatments for people with inflammatory diseases.
Dr Matt Hepworth, based at the University of Manchester’s Lydia Becker Institute of Immunology and Inflammation co-wrote this paper with Professor David Withers of the University of Birmingham.
“We wanted to understand more about how these cells decide whether to be helpful or inflammatory by looking at how transcriptional networks inside these cells regulate whether they do one thing or the other,” says Matt. “We designed in a series of studies in animal models in which we could switch off transcription factors. This enabled us to work out which pathways are important for turning them into an inflammatory cell or retaining their beneficial functions.”
“We found that there are specific requirements for the positive effects of these cells, and that some transcription factors control their ability to become inflammatory. And if you can switch them off, that reverts the ILC back to a more beneficial function.”The paper, “Reciprocal transcription factor networks govern tissue-resident ILC3 subset function and identity,” was published in Nature Immunology at the end of September.
Matt and his colleagues would now like to explore new avenues to dampen down and inhibit the hyperactivation of these cells in inflammatory disease. The research has also revealed modules of genes that may be associated with helping these cells continue their homeostatic, healthy functions.
“This work has told us a lot about how these cells function and given us ideas of areas where we could exploit that information – perhaps boosting them in certain situations, for example, where you might have an infectious disease such as an E.coli infection.”
You can find out more about Matt and his research here.
Dr Tim Blower’s recent paper in Nucleic Acids Research identifies two defence systems that protect bacteria from a wide range of phages.

The hope is that these characterised bacterial phage-defence systems could be used as biotechnological tools to recognise modified DNA and help map the human epigenome.
This paper results from a long-term collaboration between Tim, Associate Professor in the Department of Biosciences and his colleagues at Durham University, along with researchers from Northumbria and Liverpool Universities and New England Biolabs.
They reveal that there are two complementary defence systems at work to protect bacteria from viruses. The type I BREX system protected the bacteria from phages without any modifications to their DNA. Another novel system involves the BrxU restriction enzyme and provides a second line of defence by protecting bacteria from phages with modified DNA.
The open access paper, “The phage defence island of a multidrug resistant plasmid uses both BREX and type IV restriction for complementary protection from viruses,” was published in October in Nucleic Acids Research.
The team now have a detailed picture of how the BrxU system works. Its potential to be a useful research tool lies in the fact that it recognises the same DNA modifications that appear in the human genome and are altered in diseases such as cancer and neurodegeneration.
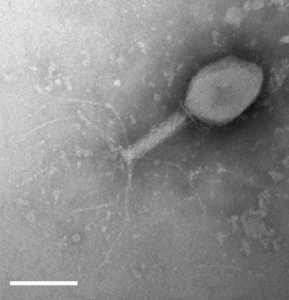
A fascinating aspect of this research is that Tim took microbiology lessons for his Durham University undergraduates out of the lecture theatre and into the local area, to gather bacteriophages from the River Wear, College ponds and other waterways around Durham. The 30 phages they isolated were then used to test against the defence systems. The paper acknowledges the 97 undergraduates involved, one of whom, Izaak Beck, went on to join Tim’s lab to do a MSc and is now in the final year of his PhD. Several others went on to study biomedical science at higher levels and now work in the biomedical sector.
Tim was awarded the Lister Prize in 2019 and we are delighted that our funding has been able to support this research.
“The flexibility of the Lister funding was key to ensure the passing-on of expertise and knowledge, and in getting the best research outputs,” says Tim.
“This work was started by a BBSRC-funded PhD student in my lab, David Picton, and through Lister funding I was able to employ him for a further year as a post-doc. During that time, this study was completed and accepted for publication, and further manuscripts are in development. Lister funding has also employed Dr Abbie Kelly as a second post-doc who is working to expand the project.”
You can read more about the Blower Lab’s research on their website.



